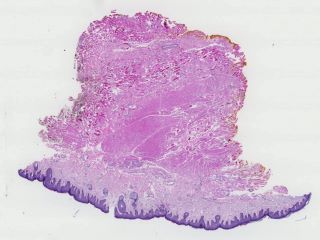
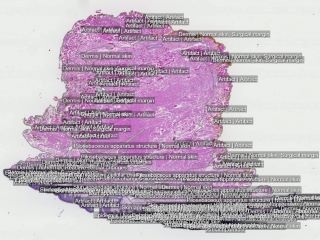
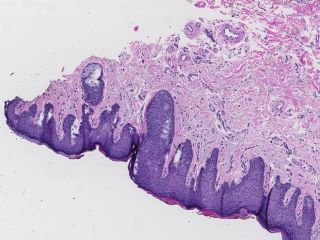
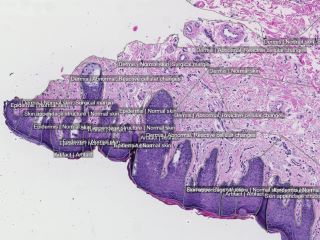
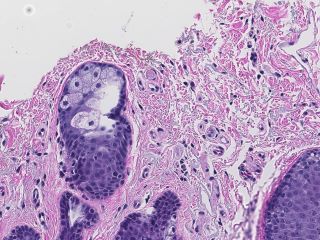
The dataset consists of 99 H&E-stained whole slide skin images (WSI) - 49 abnormal and 50 normal cases. All significant abnormal findings identified are outlined and categorized into 13 types such as actinic keratosis, basal cell carcinoma and dermatofibroma. Other tissue components, such as epidermis, adnexal structures, as well as the surgical margin are delineated to create a complete histological map. In total, 16741 separate annotations have been made to segment the different tissue structures and link them to ontological information.
Keywords: Pathology, Skin, Cancer, Whole slide imaging, Annotated.
Sample images
Sample images with reduced image quality. Please click to preview.
Dataset information
| Short name | DRSK |
|---|---|
| Origin | Clinical |
| Cite as |
Karin Lindman, Jerónimo F. Rose, Martin Lindvall, and Caroline Bivik Stadler
(2019)
Skin data from the Visual Sweden project DROID
doi:10.23698/aida/drsk [BibTeX format] |
| Field | Pathology |
| Organ |
Skin |
| Age span | 20-93 years |
| Title | Skin data from the Visual Sweden project DROID |
| Author |
Karin Lindman
Jerónimo F. Rose Martin Lindvall Caroline Bivik Stadler |
| Year | 2019 |
| DOI | doi:10.23698/aida/drsk |
| Status | Completed |
| Version | 1.1.0 |
| Scans | 99 |
| Annotations | 16741 |
| Size | 32.36GB |
| Resolution | 20X single plane |
| Modality |
SM
|
| Scanner |
Scanscope AT (Aperio, US) NanoZoomer XR (Hamamatsu, Japan) NanoZoomer XRL (Hamamatsu, Japan) |
| Stain | H&E (hematoxylin and eosin) |
| Phase | |
| References | |
| Copyright | Copyright 2019 Linköping University, Claes Lundström |
| Access |
Available under the following licenses, described in the License section below.
Controlled access
AIDA BY license
|
Annotation
One physician was responsible for the manual annotations controlled by a second pathologist. Accurate annotations were made over the whole tissues. 16741 separate annotations were made.
The following findings were annotated in the abnormal cases: actinic keratosis, basal cell carcinoma, dermatofibroma, dysplastic nevus, intradermal nevus, keratoachantoma, lentigo malignant melanoma, malignant melanoma, malignant melanoma in situ, scar, seborrheic keratosis, squamous cell carcinoma and squamous cell carcinoma in situ. Other areas annotated: abnormal, acanthosis, artifacts, dermis, epidermis, fibrosis, fibrin body, granuloma, inflammation, inflammatory edema, normal, perichondrium, reactive cellular changes, skin appendage structure, surgical margins, structure of cartilage of auditory canal, subcutaneous fatty tissue, subcutaneous tissue, surgical margins and from which body part the skin was excised.
In the normal skin cases were following structures chosen to be annotated: artifact, dermis, epidermis, normal skin, perichondrium, skin and subcutaneous structure, skin appendage structure, skin structure, structure of cartilage of auditory canal, subcutaneous fatty tissue, subcutaneous tissue and surgical margins.
File formats
Pixel position scaling
Coordinates given are relative to the image width. To get the correct pixel position, X coordinates (and Y coordinates!) should therefore be multiplied with the image width.
License
Controlled access
Free for use in legal and ethical medical diagnostics research.
To request access to the dataset, use the Apply for access button below.
Note that the recipient researcher must hold at least a PhD degree in a
relevant field and that the applicant should be an authorized signatory who can
legally enter into data sharing agreements on the behalf of the institution.
Help with applying for controlled access.
AIDA BY license
Copyright 2019 Linköping University, Claes Lundström
Permission to use, copy, modify, and/or distribute this data within Analytic Imaging Diagnostics Arena (AIDA) for the purpose of medical diagnostics research with or without fee is hereby granted, provided that the above copyright notice and this permission notice appear in all copies, and that publications resulting from the use of this data cite the following works:
Karin Lindman, Jerónimo F. Rose, Martin Lindvall, and Caroline Bivik Stadler (2019) Skin data from the Visual Sweden project DROID doi:.
THE DATA IS PROVIDED “AS IS” AND THE AUTHOR DISCLAIMS ALL WARRANTIES WITH REGARD TO THIS DATA INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS. IN NO EVENT SHALL THE AUTHOR BE LIABLE FOR ANY SPECIAL, DIRECT, INDIRECT, OR CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR CHARACTERISTICS OF THIS DATA.