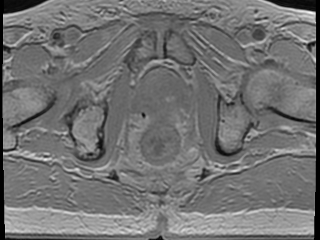
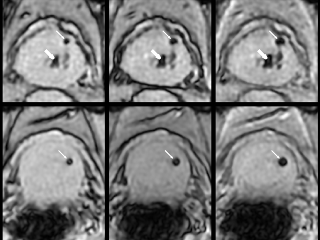
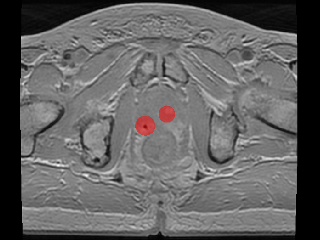
Multi-echo gradient echo (MEGRE) MRI data with 8 different echo times (2.38-23.6 ms) from 326 + 40 prostate cancer patients with gold fiducial markers inserted into the prostate (train/validation + test dataset). A scientific paper that utilizes this dataset for deep learning has been published. Underlying description of the technique and its first use has been described in a previous publication.
The data contains an image volume for each patient, for each echo time. The center of mass from the three inserted prostate gold fiducial markers was manually defined. The ground truth label for this dataset consist of spherical objects with a radius of 1-12 mm, inserted in the center of mass defined locations. Method in the paper uses 9 mm radius.
Keywords: Gold fiducial marker, MRI only, Radiotherapy, MRI radiation therapy, Prostate, Cancer, Radiology, Annotated.
Sample images
Sample images with reduced image quality. Please click to preview.
Dataset information
| Short name | MEGRE |
|---|---|
| Origin | Clinical |
| Cite as |
Christian Jamtheim Gustafsson
(2020)
Multi-echo gradient echo (MEGRE)
doi:10.23698/aida/megre [BibTeX format] |
| Field | Radiology |
| Organ |
Prostate |
| Age span | 49-85 |
| Title | Multi-echo gradient echo (MEGRE) |
| Author |
Christian Jamtheim Gustafsson
|
| Year | 2020 |
| DOI | doi:10.23698/aida/megre |
| Status | Completed |
| Version | 1.2.0 |
| Scans | 366 |
| Annotations | 692 |
| Size | 160.31GB |
| Resolution | Scan resolution 2.8x1.46x1.46 mm, reconstructed to 0.47x0.47x2.8 mm. Each patient has 28-34 slices with 512x512 image matrix. |
| Modality |
MR
|
| Scanner |
GE Discovery 750w 3T |
| Stain | |
| Phase | |
| References |
|
| Copyright | Copyright 2020 Skånes Universitetssjukhus, Christian Jamtheim Gustafsson |
| Access |
Available under the following licenses, described in the License section below.
Controlled access
AIDA BY license
|
Annotation
For 100 out of the 326 patients in the train/validation dataset the MEGRE images were annotated in MATLAB where the center of mass (CoM) coordinates of the gold fiducial marker signal voids were defined by one observer. For the remaining 226 of the patients in the train/validation dataset and the 40 patients in the test dataset MEGRE images, echo number one or two was exported to the treatment planning system Eclipse v.15.1 where the CoM points of the gold fiducial marker signal voids were defined by multiple observers and exported as DICOM RT-structures. Further details on this are provided in an article currently in review for publication. All fiducial identifications were confirmed to be correct using the corresponding CT images.
Kernels
See github repository for pre-processing and method source code
https://github.com/jamtheim/MEGRE-Net
File formats
Compressed NIfTI (.nii.gz) file format for both images and ground truth segmentations. The data contains an image volume for each patient, for each echo time.
Data augmentation was performed for each of the 326 patients with random image rotations in an interval between -15 to 15 degrees around the superior-inferior patient axis using linear interpolation. Image data for each echo for each patient time was handled separately but subjected to the same amount of rotation. The same rotation was applied to the ground truth labels using nearest neighbor interpolation (to avoid producing non-binary mask values). The augmented data and labels for each patient data was saved as a new subject, producing a total of 652 subjects in the train/validation dataset, which equaled a total of 1946 fiducial objects. No data augmentation was performed for the test data set containing 40 patients.
All image data for all echos in all datasets was independently N4 bias field corrected in an image pre-processing pipeline.
Train/validation data set and ground truth label:
XXXXXXXXX = Real patients
XXXXXXXXXAug = Augmented data
Test data set and ground truth label:
XXXXXXXXXtest= real patients
One patient had four fiducial markers inserted and was excluded in the deep learning study (in review). This patient has suffix _4fid in its name. Each patient volume for each echo in test data set was subjected to Z-score normalization after N4 bias field correction.
Folder names:
Patients40processedN4v2Normalized for Test Data
Patients326processedPooledAugN4v2 for Training/Validation Data
Folder structure for image files:
echo_1/ XXXXXXXXX_echo-1.nii.gz
echo_2/ XXXXXXXXX_echo-2.nii.gz
…
echo_8/ XXXXXXXXX_echo-8-nii.gz
Folder structure for ground truth files:
GT1/ XXXXXXXXX_coords.nii.gz
GT2/ XXXXXXXXX_coords.nii.gz
…
License
Controlled access
Free for use in legal and ethical medical diagnostics research. Please contact the dataset provider for terms of access.
You are invited to send an access request email from your institutional account.
Clicking the access request email link above should open a draft email message in a new window, to help you provide relevant information for efficient request evaluation. If the above link does not work for you, then please click to view the suggested email text.
cc: aida-data@nbis.se
Subject: Requesting access to dataset doi:10.23698/aida/megre
Hi!
I work at INSTITUTION in COUNTRY, emailing from my institutional account.
I would like to access the dataset doi:10.23698/aida/megre, for use in ethical and legal medical diagnostics research. Could you please send me a data sharing agreement template?
Our planned use of the data can be summarized as:
BRIEF_DESCRIPTION_OF_PLANNED_ACTIVITIES
We are aware that publications resulting from the use of this data must include indicated authors of this dataset in the author list.
Dataset: https://datahub.aida.scilifelab.se/10.23698/aida/megre
Example agreement template: https://datahub.aida.scilifelab.se/sharing/templates/
Template placeholders:
Research PI ("Recipient Scientist"):
Name: PI_NAME (cc here)
Title: PI_TITLE (ph d or better, in relevant field)
Name of institution: INSTITUTION_NAME
Name of department: DEPARTMENT_NAME
Institution postal address: POSTAL_ADDRESS
Authorized signatory (if other than research PI):
Name: SIGNATORY_NAME (cc here)
Title: SIGNATORY_TITLE
The Research PI has a PhD degree or better in a relevant field, and their institutional email address is in CC in this email conversation. The Authorized signatory is the person who signs this type of legal agreement on behalf of this institution, and their institutional email address is in CC in this conversation.
/MY_NAME
Click to create draft email (new window). Use your institutional account.
AIDA BY CA license
Copyright 2020 Skånes Universitetssjukhus, Christian Jamtheim Gustafsson
Permission to use, copy, modify, and/or distribute this data within Analytic Imaging Diagnostics Arena (AIDA) for the purpose of medical diagnostics research with or without fee is hereby granted, provided that the above copyright notice and this permission notice appear in all copies, and that publications resulting from the use of this data cite the following works:
Christian Jamtheim Gustafsson (2020) Multi-echo gradient echo (MEGRE) doi:10.23698/aida/megre.
THE DATA IS PROVIDED “AS IS” AND THE AUTHOR DISCLAIMS ALL WARRANTIES WITH REGARD TO THIS DATA INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS. IN NO EVENT SHALL THE AUTHOR BE LIABLE FOR ANY SPECIAL, DIRECT, INDIRECT, OR CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR CHARACTERISTICS OF THIS DATA.