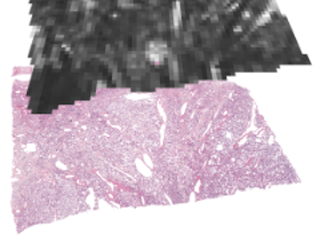
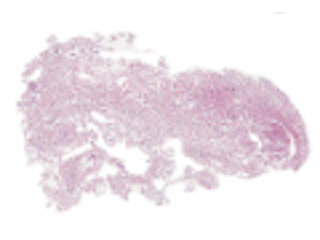
Mean diffusivity (MD) and fractional anisotropy (FA) obtained with diffusion tensor imaging (DTI) have been associated with cell density and tissue anisotropy across tumors, but these associations have been challenged at the microscopic level and several additional histological features have been suggested as contributing to MD and FA. To facilitate investigation of the biological underpinnings of DTI parameters, we performed ex-vivo dMRI at 200 μm isotropic resolution on 16 excised meningioma tumor samples. The samples together span a variety of microstructural features: six different meningioma types and two different grades. Diffusion tensor imaging (DTI) was used to produce maps such as MD, FA, in-plane FA (FAIP), axial diffusivity (AD) or radial diffusivity (RD). The maps were coregistered to H&E (hematoxylin & eosin) and VEGF-stained histological slides.
In this repository, we provide raw and analysed DTI maps coregistered to H&E- and VEGF-stained histology slides, as well as an example analysis of the data that aims to quantify the degree to which cell density (CD), structure anisotropy (SA), as determined from histology, in comparison with convolutional neural network (CNN) account for the intra-tumor variability of MD and FAIP in meningioma tumors. The pipeline used to process the raw DTI data and the coregistration tools are hosted by GitHub and the code related to the our example analysis are available here. Please refer and cite our two journal articles mentioned in the section References below for further information on the processing and if you find this data useful. We hope that data can be used in research and education concerning the link between the meningioma microstructure and parameters obtained by diffusion MRI.
Keywords: Radiology, Microstructural features, Mean diffusivity, Fractional anisotropy, cCell density, Cellularity, Meningioma, Coregistration, Hematoxylin & eosin, VEGF.
Sample images
Sample images with reduced image quality. Please click to preview.
Dataset information
| Short name | MICROMEN |
|---|---|
| Origin | Clinical |
| Cite as |
Jan Brabec, Elisabet Englund, Johan Bengzon, Filip Szczepankiewicz, Danielle van Westen, Pia C Sundgren, and Markus Nilsson
(2023)
Coregistered H&E- and VEGF-stained histology slides with diffusion tensor imaging data at 200 μm resolution in meningioma tumors
doi:10.23698/aida/micromen [BibTeX format] |
| Field | Radiology |
| Organ |
Brain |
| Age span | |
| Title | Coregistered H&E- and VEGF-stained histology slides with diffusion tensor imaging data at 200 μm resolution in meningioma tumors |
| Author |
Jan Brabec
Elisabet Englund Johan Bengzon Filip Szczepankiewicz Danielle van Westen Pia C Sundgren Markus Nilsson |
| Year | 2023 |
| DOI | doi:10.23698/aida/micromen |
| Status | Completed |
| Version | 1.0.0 |
| Scans | 16 |
| Annotations | 0 |
| Size | 390.06GB |
| Resolution | Histology slides 0.5 µm ⤫ 0.5 µm. DTI images: 200 µm x 200 µm x 200 µm. |
| Modality |
MR
|
| Scanner |
Bruker 9.4 T BioSpec Avance III MRI scanner Microscope slide scanner Hamamatsu NanoZoomer S360 The data processed with an in-house DTI pipeline and registration tool available at repositories listed in Kernels |
| Stain | Hematoxylin & Eosin (H&E) and VEGF |
| Phase | |
| References |
|
| Copyright | Copyright 2023 Lund University, Markus Nilsson |
| Access |
Available under the following licenses, described in the License section below.
Controlled access
AIDA BY license
|
Annotation
Histology slides coregistered by landmark-based approach with DTI images of meningioma tumor samples.
File formats
Histology slides: .tif and .mat files. DTI images: .nii and .mat files.
Kernels
The pipeline used to process the raw DTI data and the coregistration tools are available at https://github.com/jan-brabec/microimaging_histology_DIB
The code related to the our example analysis are available at https://github.com/jan-brabec/microimaging_vs_histology_in_meningeomas.
File formats
Pixel position scaling
Coordinates given are relative to the image width. To get the correct pixel position, X coordinates (and Y coordinates!) should therefore be multiplied with the image width.
License
Controlled access
Free for use in legal and ethical medical diagnostics research. Please contact the dataset provider for terms of access.
You are invited to send an access request email from your institutional account.
Clicking the access request email link above should open a draft email message in a new window, to help you provide relevant information for efficient request evaluation. If the above link does not work for you, then please click to view the suggested email text.
cc: markus.nilsson@med.lu.se,aida-data@nbis.se
Subject: Requesting access to dataset doi:10.23698/aida/micromen
Hi!
I work at INSTITUTION in COUNTRY, emailing from my institutional account.
I would like to access the dataset doi:10.23698/aida/micromen, for use in ethical and legal medical diagnostics research. Could you please send me a data sharing agreement template?
Our planned use of the data can be summarized as:
BRIEF_DESCRIPTION_OF_PLANNED_ACTIVITIES
Dataset: https://datahub.aida.scilifelab.se/10.23698/aida/micromen
Example agreement template: https://datahub.aida.scilifelab.se/sharing/templates/
Template placeholders:
Research PI ("Recipient Scientist"):
Name: PI_NAME (cc here)
Title: PI_TITLE (ph d or better, in relevant field)
Name of institution: INSTITUTION_NAME
Name of department: DEPARTMENT_NAME
Institution postal address: POSTAL_ADDRESS
Authorized signatory (if other than research PI):
Name: SIGNATORY_NAME (cc here)
Title: SIGNATORY_TITLE
The Research PI has a PhD degree or better in a relevant field, and their institutional email address is in CC in this email conversation. The Authorized signatory is the person who signs this type of legal agreement on behalf of this institution, and their institutional email address is in CC in this conversation.
/MY_NAME
Click to create draft email (new window). Use your institutional account.
AIDA BY license
Copyright 2023 Lund University, Markus Nilsson
Permission to use, copy, modify, and/or distribute this data within Analytic Imaging Diagnostics Arena (AIDA) for the purpose of medical diagnostics research with or without fee is hereby granted, provided that the above copyright notice and this permission notice appear in all copies, and that publications resulting from the use of this data cite the following works:
Jan Brabec, Elisabet Englund, Johan Bengzon, Filip Szczepankiewicz, Danielle van Westen, Pia C Sundgren, and Markus Nilsson (2023) Coregistered H&E- and VEGF-stained histology slides with diffusion tensor imaging data at 200 μm resolution in meningioma tumors doi:10.23698/aida/micromen.
THE DATA IS PROVIDED “AS IS” AND THE AUTHOR DISCLAIMS ALL WARRANTIES WITH REGARD TO THIS DATA INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS. IN NO EVENT SHALL THE AUTHOR BE LIABLE FOR ANY SPECIAL, DIRECT, INDIRECT, OR CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR CHARACTERISTICS OF THIS DATA.