AIDA Data Hub » AIDA Data Sharing Policy
This section contains examples of what AIDA considers correct anonymization along with rationale, to support researchers trying to choose appropriate anonymization methods for their own datasets. The intention is for this document to evolve in open discussions over time, with the purpose of establishing a national common view on anonymization in medical imaging, which has the potential to greatly facilitate medical imaging research in Sweden and beyond.
Contents:
- General guidelines
- Computed Tomography Pulmonary Angiography (CTPA) data
- Skeletal data from the Visual Sweden project DROID
- Axillary lymph nodes in breast cancer cases
As we learned in the Protective measures part of the Legal discussion section, GDPR does not require absolute certainties -which is often difficult or impossible to establish- but appropriate measures relative to the risk. The AIDA stance on appropriate measures is described and motivated in the AIDA GDPR policy 1.0 section Definition of anonymous data. Essentially, this states that thoughtful anonymization is an appropriate protective measure for using clinical imaging data in research. This statement is not controversial, but supported by a veritable mountain of evidence in the form of images that are regularly and continually published openly in scientific journals as part of good clinical research practice.
What then is appropriate anonymization in medical imaging?
While we have concluded several times already (cf for example here) that this question must in each case be finally answered by the researchers themselves, we can provide a few examples of what AIDA considers correct anonymization, to support researchers trying to make thoughtful decisions regarding their own datasets.
Each case below has been deemed appropriately anonymous for AIDA Data Hub sharing, with select images published openly as examples on the AIDA dataset register.
General guidelines
While anonymization must always be assessed on a case-by-case basis, we can condense a few guidelines based on links and examples available in this section.
Associated information
Even in cases where an image is inarguably anonymous, it may still be potentially sensitive information and identifiable through its associated data, like diagnosis, lab analysis data, time stamps, patient age and sex and so on. This is discussed in the section on Informational protective measures.
These combinations of data types can be anonymous (depending on specific circumstances which must be evaluated on a case-by-case basis):
- Images.
- Images + scanner model.
Even if the caregiver might be identified by the combination of scanners. - Images + scanner model + time period.
Even if examinations were likely carried out in the interval between scanner model launch date and dataset creation date. - Images + scanner model + time period + examination type.
Even if the image data is from an identifiable request type with a well defined examination protocol. - Images + scanner model + time period + examination type + diagnosis.
Even if a low percentage may be found positive/negative in this type of examination. - Even further characterizing data.
But you must motivate your decision yourself!
Anonymization measures for clinical imaging data
In the clinical PACS, the pixel data is stored together with large amounts of related information as structured elements in the extensive and very expressive DICOM format. Many of these elements may contain identifying information. It can be nontrivial to know which need to be removed or modified in order to ensure proper anonymization. Different approaches are differently effective for different datasets, so for each case one should strive to select a method that is satisfactory in terms of anonymization while still not negatively impacting the ability to carry out the planned research. For large scale anonymization it is good practice to use automated tools to do most of the work, and to iteratively review the output and tweak the parameters, until no more identifying information is found in the results.
For example, consider whether you need to remove:
- Report: remove number, names and initials (pat and doc), HSAID.
- Images: remove scanned images, images with unknown source, or images with low intensity variance (usually these are pictures of text, which may contain identifiable information).
You may also need to modify:
- Times: initial additive timeshift, and proportional timeshifts of inter-examination interval.
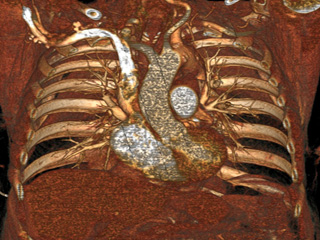
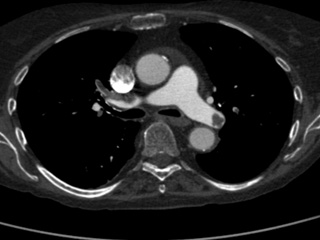
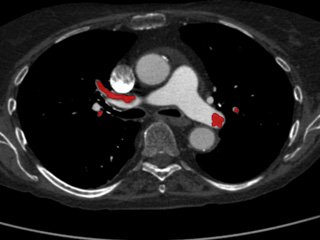
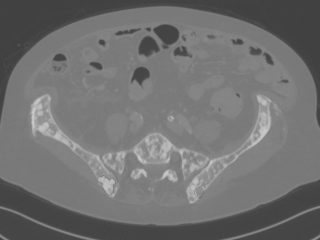
Computed Tomography Pulmonary Angiography (CTPA) data
Dataset: https://doi.org/10.23698/aida/ctpa
Example images:
30 clinical routine CTPA examinations, performed on a Philips Brilliance 64 CT or GE Lightspeed VCT. 14317 axial images (1 or 0,625mm) plus additional reformats. 5 of the CTPAs are positive for pulmonary embolism and have all the emboli carefully delineated by an experienced radiologist.
These CTPA image stacks were originally obtained in clinical routine examinations to provide care for patients. They were then extracted from the clinical PACS for research purposes and anonymized at the clinic using a programmable hardware module configured for this purpose and set up as a teleradiology destination onsite at the clinic. AIDA has developed software tools that work similarly.
In the clinical PACS, the pixel data is stored together with large amounts of related information as structured elements in the extensive and very expressive DICOM format. This data was anonymized by carefully deleting or modifying elements using automated tools, and reviewing the results afterwards for unexpected findings. Different methods are differently effective for different datasets, and several automated methods were evaluated before a method was found that was deemed adequate for this dataset, while still being feasible at scale and producing useful output for research.
For example, one of the methods that was deemed insufficient left very accurate timestamps for the examinations in the supposedly anonymized data, which together with other easily deduced properties from the dataset (eg: caregiver) and image data (eg: gender) could make reidentification by a lay person or motivated intruder plausible. In contrast, the chosen method allowed timeshifting the examinations by an arbitrary number of days, making the examinations chronologically consistent for each patient, but making their identities harder to deduce.
The fine tuning of the anonymization method nevertheless required multiple iterations of automated filtering and manual review to identify and mitigate all elements with potentially identifying information. This also had to be repeated for each scanner, as different instrument manufacturers tend to favor different types of elements for their recording of the same types of information. Notably, several elements were found to contain very accurate examination timestamps, and some images (such as the radiation dose report) contained burned-in patient information and thus could not be included in the export.
When full review of all exported DICOM data no longer revealed any plausibly identifying information for pilot cases, the configuration was deemed fit for export at scale. When exporting at scale, one examination out of every 100 were again selected for complete review of all exported DICOM data, to confirm the validity of the method, and to verify that no new types of potentially identifying information was now being added to the allowed elements (eg by clinical staff or otherwise).
It is reasonable to expect that every combination of caregiver and instrument manufacturer will have their own “house rules” for how DICOM elements are used, so it is recommended that anyone involved in anonymization carry out this type of full review of exported DICOM data, to establish which elements are safe to disclose and which aren’t, in relation to other information available on the dataset.
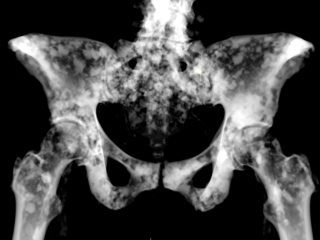
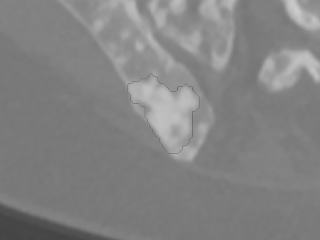
Skeletal data from the Visual Sweden project DROID
Dataset: https://doi.org/10.23698/aida/drske
Example images:
36 annotated radiology cases showing lytic and lytic/sclerotic (blastic) metastases i.e. bone regenerative and degenerative.
- Data was anonymized at the caregiver before disclosure.
- Data was collected retrospectively from routine clinical practice, representing a small part of the total number of similar examinations carried out at the caregiver.
- Data was extracted in an automatic process, where a computer program extracted the data and removed identifying information such as name, personal identification number and telephone number, or replaced it with generated information. Indirectly identifying information such as request- and examination numbers was also removed or replaced.
- Data was inspected visually by medical staff before disclosure, to ensure no identifying features were present in the data.
- This procedure was described in the (approved) research ethical review application, which also described the intent to publish and share data thusly anonymized for the purposes of research validation and as a basis for further research and AI tool development.
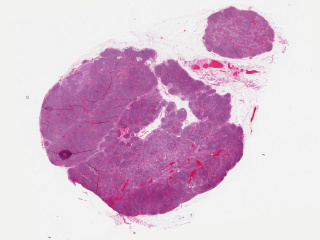
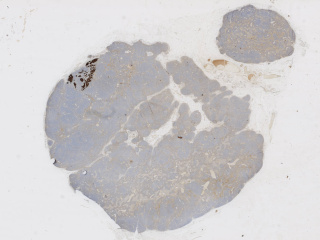
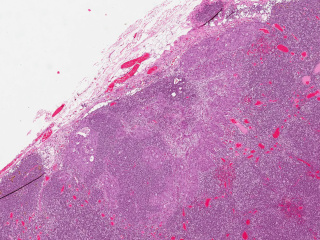
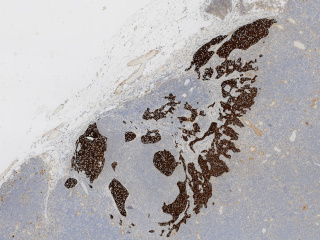
Axillary lymph nodes in breast cancer cases
Dataset: https://doi.org/10.23698/aida/brln
Example images:
Whole slide imaging of 396 full cases of axillary lymph nodes in breast cancer cases. Included are both sentinel node surgery and axillary resections pre, peri or post breast cancer surgery or treatment. Sentinel node cases are cut in three levels (stained with HE) and one additional slide immunohistochemically stained with CKAE1/AE3. The number of sentinel node cases with complete immunohistochemical staining is 321. The axillary resections are cut with one cut level as default. Included are both positive and negative cases.
- Data was anonymized at the caregiver before disclosure.
- Data was collected retrospectively from routine clinical practice, representing a small part of the total number of similar examinations carried out at the caregiver.
- Case data was extracted manually from the laboratory information system. Corresponding image data was extracted from the PACS in an automatic process, where a computer program also removed identifying information such as name, personal identification number and telephone number, or replaced it with generated information. Indirectly identifying information such as request- and examination numbers was also removed or replaced. Time stamps were removed and the order of examinations was randomized.
- Data was inspected visually by medical staff before disclosure, to ensure no identifying features were present in the data.
- This procedure was described in the (approved) research ethical review application, which also described the intent to publish and share data thusly anonymized for the purposes of research validation and as a basis for further research and AI tool development.